My research activities
My research interests lie in the field of chromatin biology and the epigenetic regulation of nuclear functions in mammals. During my PhD-thesis (2008 – 2012, ENS de Lyon, France), I studied the links between chromatin topology and DNA replication (1, 2). During my post-doc (2012 – 2016, Oxford, UK), I made use of a genetic loss-of-function screen in mouse embryonic stem cells to identify and characterize novel factors required for X-chromosome inactivation (3-5).
In September 2016, I was recruited as a Lecturer by the Paris-Sud / Paris-Saclay University and joined the Noordermeer lab (I2BC, Gif-sur-Yvette, France). I now study the contribution of the 3D-genome organization in the mono-allelic expression of genes subjected to genomic imprinting. A first preprint of our work is now hosted on BioRxiv (6).
Genomic imprinting -- Introduction
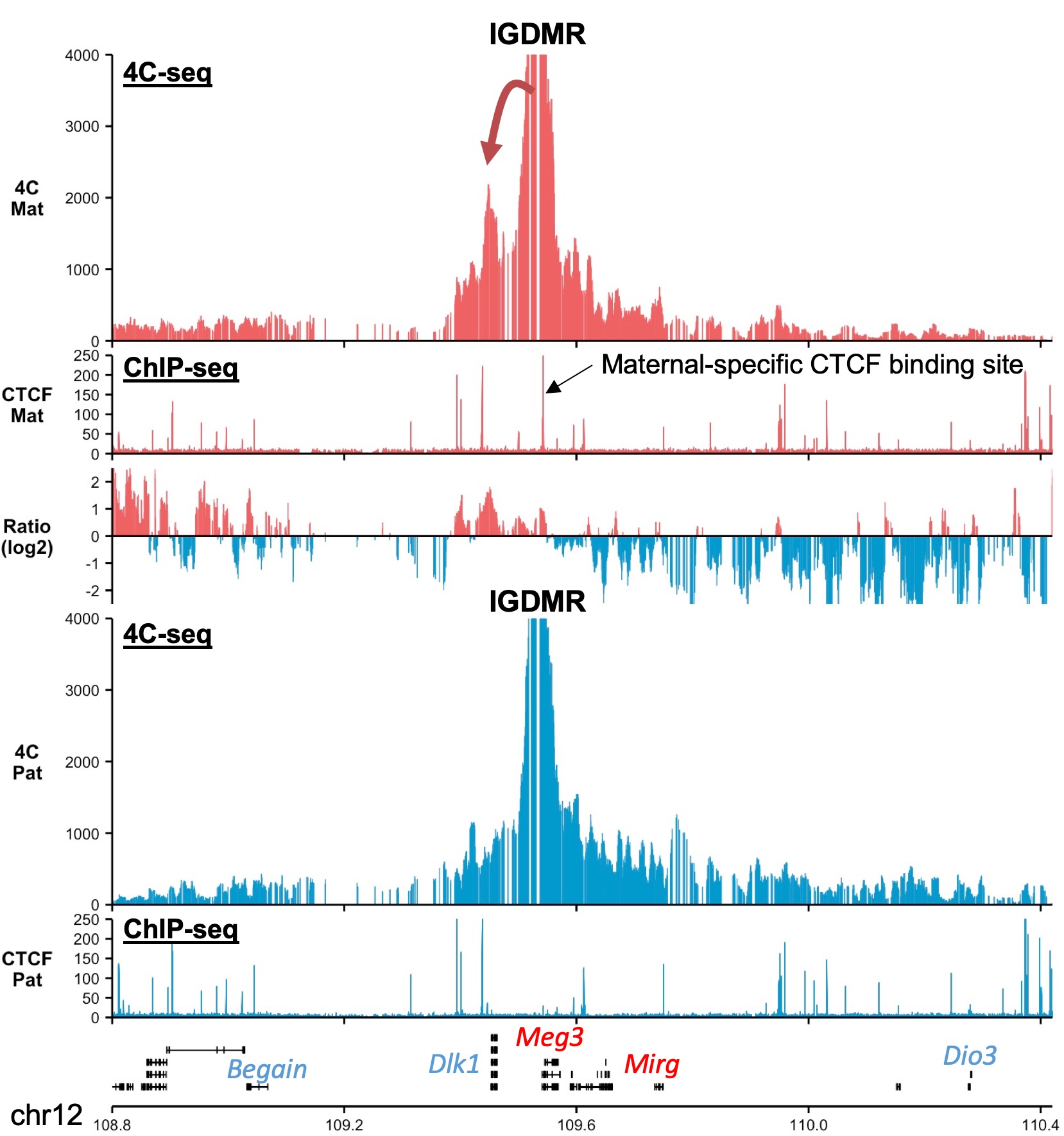
Genomic imprinting is a physiological process in mammals whereby the expression of an allele depends on its parental origin (7). The mono-allelic expression is largely determined by differentially methylation regions (DMR) that carry germline-acquired allelic DNA methylation (8). The Igf2 – H19 and Dlk1 – Dio3 domains both contain allele-specific CTCF binding site at the DMR or close to the DMR (9, 10). CTCF being one of the key 3D-genome organizers in mammals, I am combining different genomics-based approaches to comprehend the functional importance of the allelic chromatin 3D-organization for correct imprinting.
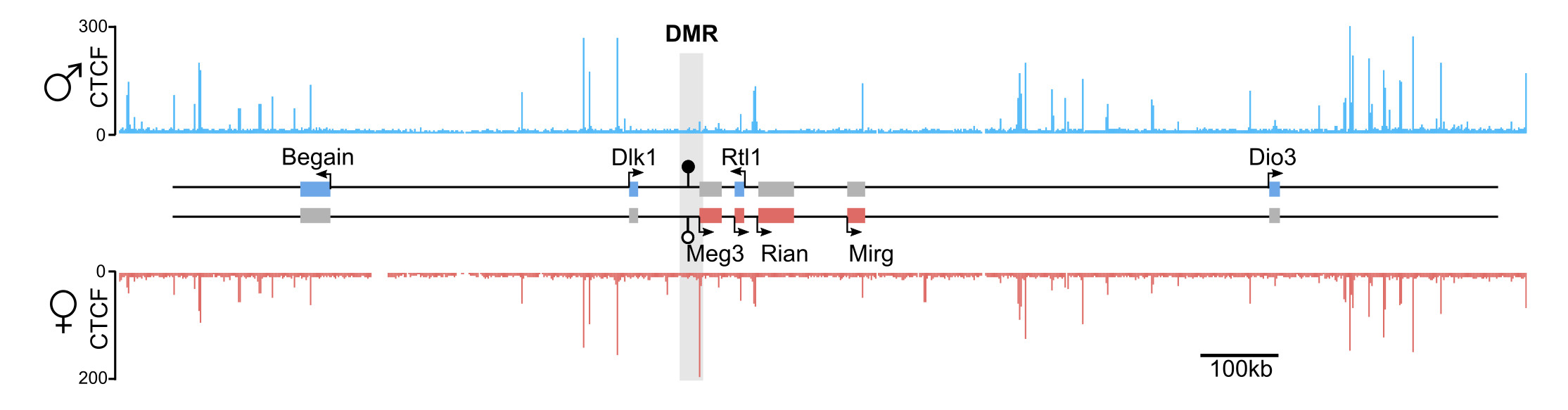
Genomic imprinting -- Our work
Using 4C-seq, our work demonstrates that the two alleles of the Igf2 – H19 and Dlk1 – Dio3 domains have pronouncedly different sub-TAD organization, due to the presence of maternal-specific CTCF binding sites that redistribute the 3D-contacts. The definite 3D-organization of the maternal allele is essential to prevent the activation of the paternally-expressed genes from the maternal chromosome. Altogether, this highlights the importance of allele-specific 3D domain organization to ensure correct mono-allelic expression. A preprint of this work, in collaboration with the lab of Robert Feil, is now hosted on BioRxiv (6).
Collaboration
This work is done in collaboration with the lab of Robert Feil.
Selected References
- 1. Baker,A., Audit,B., Chen,C.-L., Moindrot,B., Leleu,A., Guilbaud,G., Rappailles,A., Vaillant,C., Goldar,A., Mongelard,F., et al. (2012) Replication fork polarity gradients revealed by megabase-sized U-shaped replication timing domains in human cell lines. PLoS Comput Biol, 8, e1002443.
- 2. Moindrot,B., Audit,B., Klous,P., Baker,A., Thermes,C., De Laat,W., Bouvet,P., Mongelard,F. and Arneodo,A. (2012) 3D chromatin conformation correlates with replication timing and is conserved in resting cells. Nucleic Acids Res., 40, 9470–9481.
- 3. Moindrot,B., Cerase,A., Coker,H., Masui,O., Grijzenhout,A., Pintacuda,G., Schermelleh,L., Nesterova,T.B. and Brockdorff,N. (2015) A Pooled shRNA Screen Identifies Rbm15, Spen, and Wtap as Factors Required for Xist RNA-Mediated Silencing. Cell Rep, 12, 562–572.
- 4. Pintacuda,G., Wei,G., Roustan,C., Kirmizitas,B.A., Solcan,N., Cerase,A., Castello,A., Mohammed,S., Moindrot,B., Nesterova,T.B., et al. (2017) hnRNPK Recruits PCGF3/5-PRC1 to the Xist RNA B-Repeat to Establish Polycomb-Mediated Chromosomal Silencing. Mol. Cell, 68, 955–969.e10.
- 5. Nesterova,T.B., Wei,G., Coker,H., Pintacuda,G., Bowness,J.S., Zhang,T., Almeida,M., Bloechl,B., Moindrot,B., Carter,E.J., et al. (2019) Systematic allelic analysis defines the interplay of key pathways in X chromosome inactivation. Nat Commun, 10, 3129.
- 6. Llères,D., Moindrot,B., Pathak,R., Piras,V., Matelot,M., Pignard,B., Marchand,A., Poncelet,M., Perrin,A., Tellier,V., et al. (2019) CTCF controls imprinted gene activity at the mouse Dlk1-Dio3 and Igf2-H19 domains by modulating allele-specific sub-TAD structure. bioRxiv.
- 7. Ferguson-Smith,A.C. (2011) Genomic imprinting: the emergence of an epigenetic paradigm. Nat. Rev. Genet., 12, 565–575.
- 8. Kelsey,G. and Feil,R. (2013) New insights into establishment and maintenance of DNA methylation imprints in mammals. Philos. Trans. R. Soc. Lond., B, Biol. Sci., 368, 20110336.
- 9. Bell,A.C. and Felsenfeld,G. (2000) Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature, 405, 482–485.
- 10. Rosa,A.L., Wu,Y.-Q., Kwabi-Addo,B., Coveler,K.J., Reid Sutton,V. and Shaffer,L.G. (2005) Allele-specific methylation of a functional CTCF binding site upstream of MEG3 in the human imprinted domain of 14q32. Chromosome Res., 13, 809–818.


