Parametric uncertainties in Molecular Simulations
Simulation of chemical or physical systems necessitates the use of models that rely on the definitions of parameters. For example, modeling the time evolution of a chemical system in reaction requires the definition of the values of the rate constants of each possible chemical reaction. The values taken by these parameters can be obtained from experiments, inferred from calculations, or empirically determined.
Almost all parameters used in chemical or physical models are not known with extreme precision. This uncertainty in the parameter values should be taken into account when making prediction with a model, in order to give a confidence interval for that prediction. Without this, no quantitative conclusion could be made. Managing parametric uncertainty in physico-chemical models is thus a major issue.
Since my arrival in the Laboratoire de Chimie Physique in 2008, I have been working in collaboration with Dr Pascal Pernot, that developped an expertise in uncertainty management in physico-chemical models. Due to my background in molecular simulation, we decided to explore the impact of forcefield parameters uncertainty.
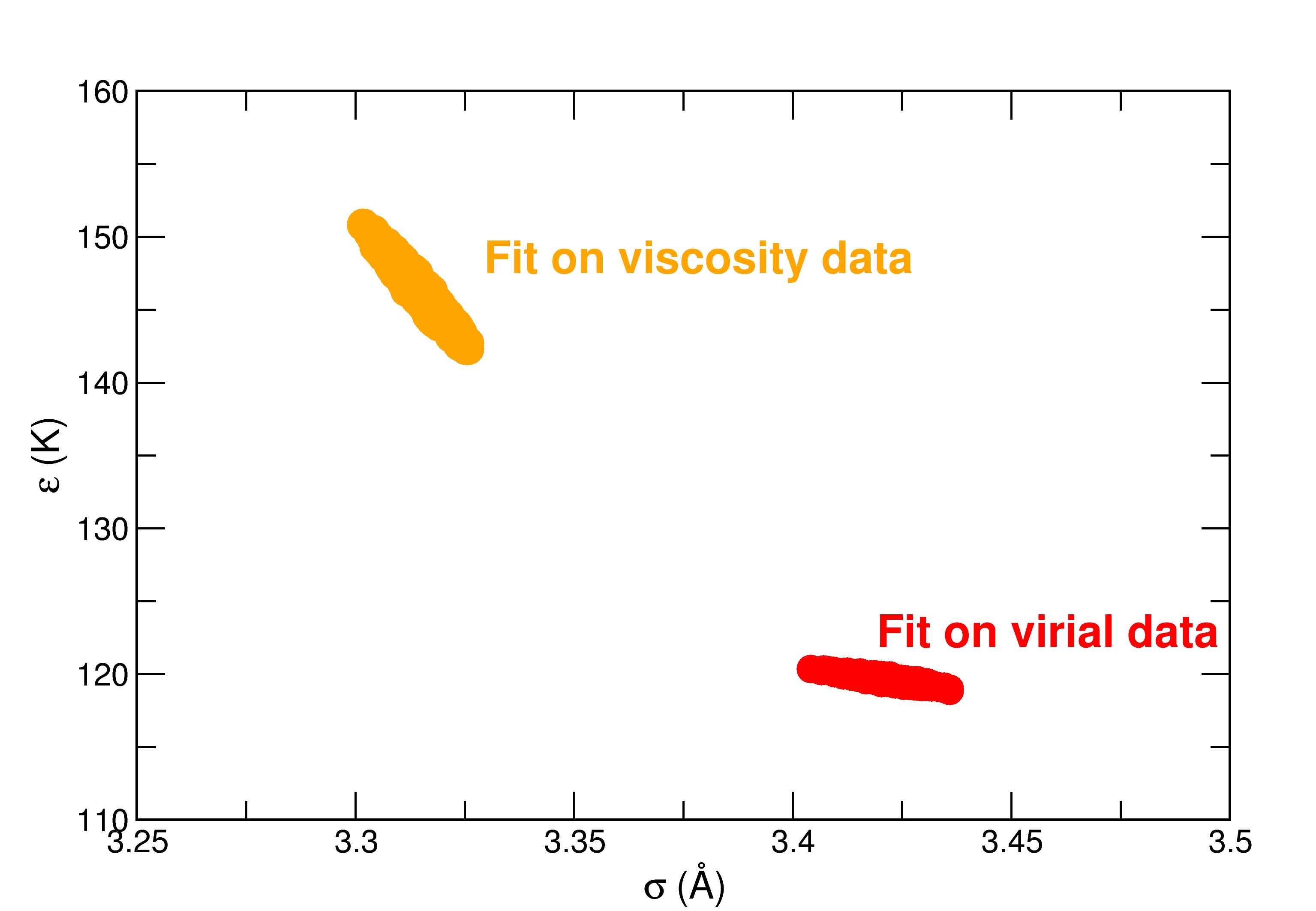
Our first work dealt with the study of a Lennard-Jones potential to simulate argon molecules. We performed a statistical calibration of the forcefield parameters in the Bayesian framework, and propagated the parameter uncertainties in molecular simulations to compute thermodynamic and transport properties of argon fluids. The main conclusiosn of this work were that:
- molecular simulations amplify parameter uncertainties
- the contribution of parameteric uncertainties in the final uncertainty budget was far from being negligible for that kind of system (being of the order of magnitude of statistical uncertainties), even if the uncertainties on the parameters were small.
 |
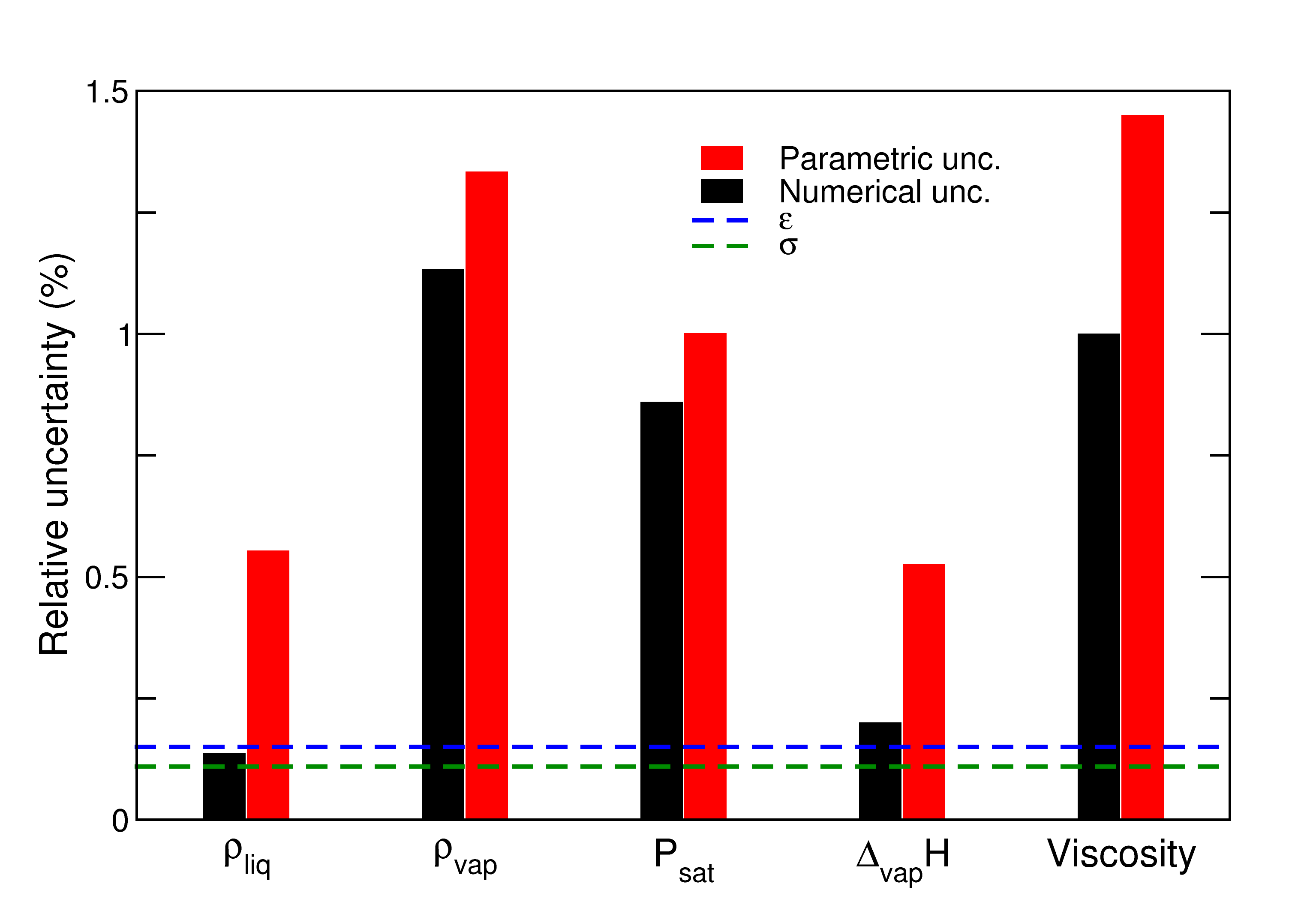 |
| Probability density functions of Lennard-Jones parameters for argon calibrated on viscosity or virial coefficients data | Total uncertainty budget for various properties of argon obtained with molecular simulation |
We are now carrying similar studies on water, modelled by a TIP4P potential, which is a 6-parameter model commonly used in molecular simulation. In that case, calibration cannot be made with the use of analytical formulas, as was the case for simple atomic Lennard-Jones potentials. Thus costly molecular simulations must be performed and exploration of the parameter space is untractable. We chose to use statistical surrogate models named gaussian processes, in order to circumvent this limitation. These tools, combined with Efficient Global Optimization methods, should be helpful in the calibration process and may help building a general framework for statistical forcefield parameters calibration.
Key words
Molecular simulation, Bayes formula, uncertainty propagation, sensitivity analysis, kriging
Publications
- "Statistical approaches to forcefield calibration and prediction uncertainty in molecular simulation". F Cailliez and P Pernot, J. Chem. Phys., 2011, 134:054124.
- "Calibration of forcefields for molecular simulation: sequential design of computer experiments for building cost-efficient kriging metamodels". F Cailliez, A Bourasseau and P Pernot, J. Comp. Chem., 2014, 35(2):130-149